SRM 2395-Human Y-Chromosome DNA Profiling Standard
NOW AVAILABLE: see http://www.nist.gov/srm
https://www-s.nist.gov/srmors/view_detail.cfm?srm=2395
The Certificate of Analysis for SRM 2395 has been updated with additional information for the 5 male components. These samples now have sequence information at over 40 different Y-STR markers and typed at an additional 3 Y-STRs and 42 Y-SNPs. This SRM may be used for calibrating methods for Y-STR typing with any primer set or commercially available kit.
[Supporting Sequence Information] [Yfiler Stutter Percentages] [Stability Information] [PCR and Sequencing Primers Used]
[Original
SRM
2395 Certificate]
Y-chromosome loci present in the following kits are covered by SRM 2395:
PowerPlex Y, Yfiler, Marligen's Signet™ Y-SNP Identification System
Variable SNPs observed in 42 Y-SNP loci examined
| SRM 2395 | AMEL | M207 | M45 | P25 | M89 | DYS391 | M2 | M170 | M172 | M201 | SNP Haplogroup |
| (A/G) | (G/A) | (C/A) | (C/T) | (C/G) | (A/G) | (A/C) | (T/G) | (G/T) | |||
| Component A | XY | G | A | A | T | C | A | A | T | G | R1b |
| Component B | XY | A | G | C | T | C | A | A | G | G | J2 |
| Component C | XY | A | G | C | C | G | G | A | T | G | E3a |
| Component D | XY | A | G | C | T | C | A | A | T | T | G |
| Component E | XY | A | G | C | T | C | A | C | T | G | I |
| Component F | XX | -- |
Original Set of Sequenced Y-STR Loci
DYS19, DYS385 a/b, DYS388, DYS389I/II, DYS390, DYS391, DYS392, DYS393, DYS426, DYS435, DYS436, DYS437, DYS438, DYS439, DYS447, DYS448, DYS460, DYS461, DYS462, GATA-H4
Additional Y-STRs with Typing Results
DYS450, DYS456, DYS458, DYS464 a/b/c/d, YCAII a/b
Added Since SRM 2395 Release
DYS635 (GATA-C4)
Component A - Male Genomic DNA 1 (23 repeats)
(TCTA)4(TGTA)2(TCTA)2(TGTA)2(TCTA)2(TGTA)2(TCTA)9
Component B – Male Genomic DNA 2 (21 repeats)
(TCTA)4(TGTA)2(TCTA)2(TGTA)2(TCTA)11
Component C – Male Genomic DNA 3 (23 repeats)
(TCTA)4(TGTA)2(TCTA)2(TGTA)2(TCTA)13
Component D – Male Genomic DNA 4 (21 repeats)
(TCTA)4(TGTA)2(TCTA)2(TGTA)2(TCTA)11
Component E – Male Genomic DNA 5 (21 repeats)
(TCTA)4(TGTA)2(TCTA)2(TGTA)2(TCTA)11
Note: We have received reports from multiple forensic DNA laboratories that elevated stutter (>20%) has been observed with the longer DYS385 allele in components B (allele 17) and C (alleles 17 and 20).
Stutter percentages for the 5 male SRM 2395 components with the 17 Yfiler loci
|
Sample Name |
A |
B |
C |
D |
E |
|
DYS456 |
9.0 |
9.4 |
10.0 |
9.2 |
9.2 |
|
DYS389I |
5.0 |
5.1 |
6.0 |
4.3 |
6.8 |
|
DYS390 |
9.5 |
7.9 |
3.4 |
5.9 |
8.2 |
|
DYS389II |
10.7 |
9.2 |
10.9 |
9.2 |
11.9 |
|
DYS458 |
7.2 |
6.5 |
8.5 |
7.2 |
7.7 |
|
DYS19 |
5.7 |
6.4 |
7.7 |
6.9 |
9.1 |
|
DYS385a |
8.4 |
8.2 |
9.8 |
8.3 |
7.4 |
|
DYS385b |
9.2 |
14.7 |
14.8 |
N/A* |
10.1 |
|
DYS393 |
9.5 |
7.4 |
8.0 |
8.5 |
8.8 |
|
DYS391 |
6.6 |
6.8 |
7.8 |
5.8 |
4.3 |
|
DYS439 |
7.5 |
5.7 |
5.5 |
5.8 |
5.0 |
|
DYS635 |
4.6 |
6.6 |
8.5 |
7.1 |
6.8 |
|
DYS392 |
12.1 |
7.5 |
8.6 |
7.1 |
8.8 |
|
DYS392 (N+3) |
6.4 |
3.4 |
3.2 |
3.1 |
3.2 |
|
Y-GATA-H4 |
7.0 |
6.6 |
6.5 |
6.5 |
6.1 |
|
DYS437 |
4.5 |
3.6 |
3.6 |
5.5 |
5.5 |
|
DYS438 |
2.5 |
1.9 |
2.1 |
2.2 |
2.5 |
|
DYS448 |
2.2 |
2.6 |
2.4 |
2.3 |
2.5 |
*N/A – stutter could not be determined due to allele in stutter position
Supporting Sequence Information
Certified allele repeat numbers using GS500-LIZ size standard for base pair sizing
|
|
DYS456 |
Repeat Motif |
bp size |
|
|
DYS607 |
Repeat Motif |
bp size |
|
A |
15 |
[AGAT]15 |
111.15 |
|
A |
19 |
[GAAG]15[GAAA][GAAG][GAAA][GAAG] |
198.16 |
|
B |
15 |
[AGAT]15 |
111.13 |
|
B |
19 |
[GAAG]15[GAAA][GAAG][GAAA][GAAG] |
197.86 |
|
C |
15 |
[AGAT]15 |
111.14 |
|
C |
19 |
[GAAG]15[GAAA][GAAG][GAAA][GAAG] |
197.94 |
|
D |
15 |
[AGAT]15 |
111.13 |
|
D |
17 |
[GAAG]13[GAAA][GAAG][GAAA][GAAG] |
190.12 |
|
E |
15 |
[AGAT]15 |
110.97 |
|
E |
18 |
[GAAG]14[GAAA][GAAG][GAAA][GAAG] |
194.02 |
|
|
|
|
|
|
|
|
|
|
|
|
DYS458 |
Repeat Motif |
bp size |
|
|
DYS635 |
Repeat Motif |
bp size |
|
A |
16 |
[GAAA]16 |
140.39 |
|
A |
23 |
[TCTA]4[TGTA]2[TCTA]2[TGTA]2[TCTA]2[TGTA]2[TCTA]9 |
260.32 |
|
B |
15 |
[GAAA]14 GGAA |
136.52 |
|
B |
21 |
[TCTA]4[TGTA]2[TCTA]2[TGTA]2[TCTA]11 |
252.66 |
|
C |
17 |
[GAAA]17 |
144.69 |
|
C |
23 |
[TCTA]4[TGTA]2[TCTA]2[TGTA]2[TCTA]13 |
260.39 |
|
D |
16 |
[GAAA]16 |
140.36 |
|
D |
21 |
[TCTA]4[TGTA]2[TCTA]2[TGTA]2[TCTA]11 |
252.17 |
|
E |
16 |
[GAAA]16 |
140.31 |
|
E |
21 |
[TCTA]4[TGTA]2[TCTA]2[TGTA]2[TCTA]11 |
252.05 |
|
|
|
|
|
|
|
|
|
|
|
|
DYS492 |
Repeat Motif |
bp size |
|
|
DYS652 |
Repeat Motif |
bp size |
|
A |
12 |
[ATT]12 |
217.81 |
|
A |
24 |
[TAATA]6 TAAAA [TAATA]10 TAAAA [TAATA]6 |
210.76 |
|
B |
12 |
[ATT]12 |
217.86 |
|
B |
25 |
[TAATA]9 TAAAA [TAATA]8 TAAAA [TAATA]6 |
215.87 |
|
C |
11 |
[ATT]11 |
214.64 |
|
C |
25 |
[TAATA]7 TAAAA [TAATA]8 TAAAA [TAATA]8 |
215.81 |
|
D |
11 |
[ATT]11 |
214.57 |
|
D |
23 |
[TAATA]6 TAAAA [TAATA]9 TAAAA [TAATA]6 |
205.64 |
|
E |
12 |
[ATT]12 |
217.67 |
|
E |
26 |
[TAATA]7 TAAAA [TAATA]11 TAAAA [TAATA]6 |
220.61 |
|
|
|
|
|
|
|
|
|
|
|
|
DYS532 |
Repeat Motif |
bp size |
|
|
DYS709 |
Repeat Motif |
bp size |
|
A |
15 |
[CTTT]15 |
490.63 |
|
A |
13 |
[CTTT]13 |
197.83 |
|
B |
11 |
[CTTT]11 |
474.47 |
|
B |
16 |
[CTTT]16 |
209.96 |
|
C |
12 |
[CTTT]12 |
478.52 |
|
C |
15 |
[CTTT]15 |
205.89 |
|
D |
15 |
[CTTT]15 |
490.38 |
|
D |
17 |
[CTTT]17 |
213.75 |
|
E |
9 |
[CTTT]9 |
466.35 |
|
E |
16 |
[CTTT]16 |
209.76 |
|
|
|
|
|
|
|
|
|
|
|
|
DYS534 |
Repeat Motif |
bp size |
|
|
DYS710 |
Repeat Motif |
bp size |
|
A |
15 |
[CTTT]15 |
214.65 |
|
A |
36 |
[AAAG]18 [AG]16 [AAAG]10 |
233.72 |
|
B |
15 |
[CTTT]15 |
214.91 |
|
B |
34.2 |
[AAAG]16 [AG]13 [AAAG]12 |
227.65 |
|
C |
15 |
[CTTT]15 |
214.87 |
|
C |
35.2 |
[AAAG]20 [AG]9 [AAAG]4 AAGG [AAAG]6 |
231.51 |
|
D |
14 |
[CTTT]14 |
210.79 |
|
D |
33.2 |
[AAAG]16 [AG]9 [AAAG]13 |
223.41 |
|
E |
14 |
[CTTT]14 |
210.81 |
|
E |
31 |
[AAAG]14 [AG]12 [AAAG]11 |
214.00 |
|
|
|
|
|
|
|
|
|
|
|
|
DYS570 |
Repeat Motif |
bp size |
|
|
DYS715 |
Repeat Motif |
bp size |
|
A |
17 |
[TTTC]17 |
264.01 |
|
A |
24 |
[TAGA]14 N20 [TGGA]10 |
195.4 |
|
B |
18 |
[TTTC]18 |
268.32 |
|
B |
21 |
[TAGA]11 N20 [TGGA]10 |
183.51 |
|
C |
18 |
[TTTC]18 |
268.21 |
|
C |
22 |
[TAGA]12 N20 [TGGA]10 |
187.36 |
|
D |
17 |
[TTTC]17 |
263.66 |
|
D |
23 |
[TAGA]13 N20 [TGGA]10 |
191.29 |
|
E |
18 |
[TTTC]18 |
267.88 |
|
E |
23 |
[TAGA]12 N20 [TGGA]11 |
191.37 |
|
|
|
|
|
|
|
|
|
|
|
|
DYS572 |
Repeat Motif |
bp size |
|
|
DYS717 |
Repeat Motif |
bp size |
|
A |
11 |
[AAAT]11 |
254.21 |
|
A |
17 |
TGTAT [TGTAC]6 [TGTAT]10 |
253.94 |
|
B |
11 |
[AAAT]11 |
254.04 |
|
B |
17 |
TGTAT [TGTAC]6 [TGTAT]10 |
253.99 |
|
C |
9 |
[AAAT]9 |
245.94 |
|
C |
17 |
TGTAT [TGTAC]6 [TGTAT]10 |
253.73 |
|
D |
9 |
[AAAT]9 |
245.87 |
|
D |
20 |
TGTAT [TGTAC]8 [TGTAT]11 |
269.00 |
|
E |
11 |
[AAAT]11 |
253.95 |
|
E |
14 |
TGTAT [TGTAC]6 [TGTAT]7 |
238.39 |
|
|
|
|
|
|
||||
|
|
DYS576 |
Repeat Motif |
bp size |
|
||||
|
A |
18 |
[AAAG]18 |
197.28 |
|
||||
|
B |
16 |
[AAAG]16 |
189.52 |
|
||||
|
C |
17 |
[AAAG]17 |
193.44 |
|
||||
|
D |
18 |
[AAAG]18 |
197.23 |
|
||||
|
E |
17 |
[AAAG]17 |
193.40 |
|
Certified and Reference Values Method Dependent (GeneScan 600-LIZ size standard used for base pair sizing)
|
|
DYS449 |
Repeat Motif |
bp size |
|
|
DYS527 |
Repeat Motif |
bp size |
|
A |
28 |
[TTTC]14N50[TTTC]14 |
349.93 |
|
A |
21 |
[GAAA]14 [GGAA]7 |
353.90 |
|
B |
32 |
[TTTC]16N50[TTTC]16 |
365.58 |
|
|
23 |
[GAAA]16 [GGAA]7 |
361.74 |
|
C |
30 |
[TTTC]16N50[TTTC]14 |
357.78 |
|
B |
22 |
[GAAA]16 [GGAA]6 |
357.71 |
|
D |
28 |
[TTTC]15N50[TTTC]13 |
349.94 |
|
C |
17* |
|
338.15 |
|
E |
27 |
[TTTC]13N50[TTTC]14 |
346.04 |
|
|
20 |
[GAAA]13 [GGAA]7 |
350.01 |
|
|
|
|
|
|
D |
15 |
[GAAA]9 [GGAA]6 |
330.41 |
|
|
DYS481 |
Repeat Motif |
bp size |
|
|
21 |
[GAAA]15 [GGAA]6 |
353.81 |
|
A |
22 |
[CTT]22 |
132.16 |
|
E |
22 |
[GAAA]15 [GGAA]7 |
357.78 |
|
B |
23 |
[CTT]23 |
135.11 |
|
|
23* |
|
361.73 |
|
C |
28 |
[CTT]28 |
150.47 |
|
|
|
|
|
|
D |
23 |
[CTT]23 |
135.15 |
|
|
DYS650 |
Repeat Motif |
bp size |
|
E |
28 |
[CTT]28 |
150.45 |
|
A |
18 |
[AAGG]17 AAAG |
283.41 |
|
|
|
|
|
|
B |
18 |
[AAGG]15 AAGA [AAAG]2 |
283.25 |
|
|
DYS522 |
Repeat Motif |
bp size |
|
C |
16 |
[AAGG]13 AAGA [AAAG]2 |
275.59 |
|
A |
10 |
[GATA]10 |
357.03 |
|
D |
18 |
[AAGG]15 AAGA [AAAG]2 |
283.27 |
|
B |
11 |
[GATA]11 |
361.02 |
|
E |
24* |
|
306.62 |
|
C |
10 |
[GATA]10 |
356.96 |
|
|
|
|
|
|
D |
12 |
[GATA]12 |
365.00 |
|
|
DYS712 |
Repeat Motif |
bp size |
|
E |
12 |
[GATA]12 |
365.16 |
|
A |
23 |
[AGAT]18 [AGAC]5 |
177.49 |
|
|
|
|
|
|
B |
22.3 |
[AGAT]2 GAT[AGAT]14[AGAC]6 |
176.42 |
|
|
|
|
|
|
C |
21 |
[AGAT]15 [AGAC]6 |
169.34 |
|
|
|
|
|
|
D |
26 |
[AGAT]17 [AGAC]9 |
189.48 |
|
|
|
|
|
|
E |
19 |
[AGAT]12 [AGAC]7 |
161.15 |
*Alleles in italic and bold are reference values because no sequence data has been generated, therefore no repeat motif is listed
PCR primer sequences for the 21 Y-STR loci of interest.
Yfiler kit primers are proprietary to Applied Biosystems
|
PCR primers |
|
|
Locus ID |
Primer Sequence |
|
DYS449-F |
CCTGGAAGTGGAGTTTGCTGT |
|
DYS449-R |
gtttcttTGGAGTCTCTCAAGCCTGTTCTA |
|
DYS456-F |
Yfiler kit (Applied Biosystems) |
|
DYS456-R |
Yfiler kit (Applied Biosystems) |
|
DYS458-F |
Yfiler kit (Applied Biosystems) |
|
DYS458-R |
Yfiler kit (Applied Biosystems) |
|
DYS481-F |
AGGAATGTGGCTAACGCTGT |
|
DYS481-R |
gtttcttACAGCTCACCAGAAGGTTGC |
|
DYS492-F |
AGATGAGCCAGGCTTCAGAC |
|
DYS492-R |
gtttcttAGTAGGGGTCAGGCACAATG |
|
DYS522-F |
CCTTTGAAATCATTCATAATGC |
|
DYS522-R |
gtttcttTCATAAACAGAGGGTTCTGG |
|
DYS527-F |
TCGCAAACATAGCACTTCAG |
|
DYS527-R |
gtttcttTTCTAGGAAGATTAGCCACAACA |
|
DYS532-F |
TTGGTTTTATGCCTTTCACT |
|
DYS532-R |
gtttcttTAGGTGACAGAGCAGGATTC |
|
DYS534-F |
CATCTACCCAACATCCATCTA |
|
DYS534-R |
gtttcttGACAAAGATGTTAGATGAATAGACA |
|
DYS570-F |
GAACTGTCTACAATGGCTCACG |
|
DYS570-R |
gtttcttTCAGCATAGTCAAGAAACCAGACA |
|
DYS572-F |
CTAAGGACGCCTCCCATACA |
|
DYS572-R |
gtttcttCTCATTCCCTATGGTTTGCAC |
|
DYS576-F |
TTGGGCTGAGGAGTTCAATC |
|
DYS576-R |
gtttcttGGCAGTCTCATTTCCTGGAG |
|
DYS607-F |
AGCATACAGCGTAATCACAGC |
|
DYS607-R |
gtttcttTCAGACAAAGCCCAGTTGAG |
|
DYS635-F |
Yfiler kit (Applied Biosystems) |
|
DYS635-R |
Yfiler kit (Applied Biosystems) |
|
DYS650-F |
TCACATGCCTACAGTCACAGC |
|
DYS650-R |
gtttcttCTCTTTCTCCCTTCCCACCT |
|
DYS652-F |
CACAGCATGGCTTGGTTTTA |
|
DYS652-R |
gtttcttTTTGCGTTATCTCTGCCTTTC |
|
DYS709-F |
GTTGCCATGGTTTCTTGCTT |
|
DYS709-R |
gtttcttCGAACCTGCAAATTGTTCAC |
|
DYS710-F |
GAGGTCAAGGCTGCAAGAATCTATGA |
|
DYS710-R |
gtttcttCATACTCTCTCCCTCCCTCTCTTTTTTC |
|
DYS712-F |
CAAGAACAGCCTGGGTAACAGTG |
|
DYS712-R |
gtttcttTTATATGGTACAGCCCATGAACACTT |
|
DYS715-F |
ATGGTTGGAAGAAAGCATTGATGA |
|
DYS715-R |
gtttcttCTAGGTAATTAGCTACCTAGTTAG |
|
DYS717-F |
GGCCGAGAGAATGGAATTGAT |
|
DYS717-R |
gtttcttCCCGAACTTCAGCACTATGAAATG |
Sequencing primer sequences for the 21 new Y-STR loci
|
Sequencing primers |
|
|
|
|
Locus ID |
Primer Sequence |
Length |
Amp size (bp) |
|
DYS449-F |
tgctgtatgttctaatcttaggttgc |
26 |
478 |
|
DYS449-R |
ctgaggccggataattgct |
19 |
|
|
DYS456-F |
actcggactggctcatcttg |
20 |
197 |
|
DYS456-R |
cccatcaactcagcccaaaac |
21 |
|
|
DYS458-F |
gggtggtggaggttactgtg |
20 |
320 |
|
DYS458-R |
ctagaggttcctgccaccac |
20 |
|
|
DYS481-F |
ttctgtgagagtgttgcgaga |
21 |
243 |
|
DYS481-R |
acccaagaagagccacacag |
20 |
|
|
DYS492-F |
tgtgcagtggacaggaagaa |
20 |
394 |
|
DYS492-R |
tgaacctgggcaacatagtg |
20 |
|
|
DYS522-F |
ccacaatgctttccttacacc |
21 |
472 |
|
DYS522-R |
agggtggggaactttctgat |
20 |
|
|
DYS527-F |
gctggaattcagaaggtgga |
20 |
543 |
|
DYS527-R |
cattttctcaaaatccagtgaga |
23 |
|
|
DYS532-F |
tgtagataatgggcaaaatgg |
21 |
589 |
|
DYS532-R |
attgcttgaacttgggatgg |
20 |
|
|
DYS534-F |
tcattgtctaccattcactttctttc |
26 |
468 |
|
DYS534-R |
tcatgatcagttcttaactcaacca |
25 |
|
|
DYS570-F |
tcatccaattgctgtctttttg |
22 |
372 |
|
DYS570-R |
gaagggcttctaagggatgc |
20 |
|
|
DYS572-F |
tgggtattgcattcaccaac |
20 |
385 |
|
DYS572-R |
ttgctttctcagatgctggtt |
21 |
|
|
DYS576-F |
tgagagactgaggtaggaagatcc |
24 |
248 |
|
DYS576-R |
tgggtgggaacatagtcaaa |
20 |
|
|
DYS607-F |
gtctcactctgtcccccaag |
20 |
244 |
|
DYS607-R |
ttcaggtccagtcatgagaaaa |
22 |
|
|
DYS635-F |
gcatgaataacgaactcactatagg |
25 |
363 |
|
DYS635-R |
gaccaaggctccatctcaaa |
20 |
|
|
DYS650-F |
tcacatgcctacagtcacagc |
21 |
280 |
|
DYS650-R |
ctctttctcccttcccacct |
20 |
|
|
DYS652-F |
aggacatgcctgtgctacaa |
20 |
300 |
|
DYS652-R |
gggaggggaagtacatggaa |
20 |
|
|
DYS709-F |
tttttgcctgccatattggt |
20 |
293 |
|
DYS709-R |
tgacgagttaatgggtgcag |
20 |
|
|
DYS710-F |
gcagtcccagctactccaaa |
20 |
297 |
|
DYS710-R |
catactccctccctcccttc |
20 |
|
|
DYS712-F |
gattgcttgagcccagaagt |
20 |
248 |
|
DYS712-R |
gccaccagggattatgatatg |
21 |
|
|
DYS715-F |
tgatagatggataaatggatgaatg |
25 |
333 |
|
DYS715-R |
tctatctcatctttgtctctttctgc |
26 |
|
|
DYS717-F |
gccatggatggtggtagaac |
20 |
339 |
|
DYS717-R |
ttgttcaggtgaggaatatgct |
22 |
|
Results from the multiple analyses of the original two units of SRM 2395 obtained from MSD are averaged together.
DNA concentrations of SRM 2395 components A – F as determined by multiple qPCR analysis runs.
IH unit stored in-house since production
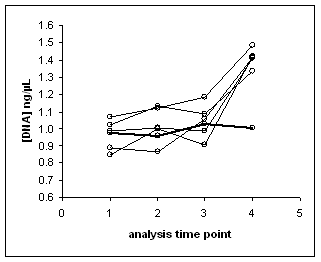
MSD units stored at MSD: 2 units obtained from MSD, n=4 is the average of replicates from each unit. The trends for the results from the two MSD units are displayed in figure below.
|
|
-20 °C IH |
-20 °C MSD |
-20 °C MSD |
-20 °C MSD |
-20 °C MSD |
|||||
|
|
n=2 10/30/07 |
n=4 10/25/07 AM |
n=4 10/25/07 PM |
n=4 10/30/07 |
n=0,1, or2 11/29/07 |
|||||
|
Component |
Avg |
sd |
Avg |
sd |
Avg |
sd |
Avg |
sd |
Avg |
sd |
|
A |
2.06 |
0.35 |
0.89 |
0.13 |
0.87 |
0.16 |
1.05 |
0.11 |
1.41 |
0.09 |
|
B |
1.59 |
0.42 |
0.85 |
0.10 |
1.00 |
0.13 |
0.91 |
0.20 |
1.42 |
0.01 |
|
C |
1.66 |
0.42 |
1.07 |
0.18 |
1.12 |
0.14 |
1.19 |
0.14 |
1.49 |
0.06 |
|
D |
1.43 |
0.40 |
0.99 |
0.21 |
1.01 |
0.23 |
0.99 |
0.17 |
1.41 |
0.15 |
|
E |
1.55 |
0.47 |
1.02 |
0.11 |
1.13 |
0.16 |
1.09 |
0.15 |
1.33 |
0.06 |
|
F |
1.36 |
0.14 |
0.97 |
0.04 |
0.96 |
0.11 |
1.03 |
0.13 |
1.01 |
0.07 |
Trends of the analyses of the two MSD units of SRM 2395. Each line represents a component of SRM 2395, points along the lines correspond to analyses DNA concentrations. Components A-E are approaching the original nominal DNA concentration of 2 ng/µL. The DNA concentration of component F appears constant at 1 ng/µL.
Return to Y-Chromosome Section of STRBase
Last Updated: 12/15/2009