SRM 2372-Human DNA Quantitation Standard
NOW AVAILABLE: see http://www.nist.gov/srm
https://www-s.nist.gov/srmors/view_detail.cfm?srm=2372
Additional Information:
SRM 2372 Human DNA Quantitation Standard consists of three well-characterized human genomic DNA materials solubilized in 10 mmol/L 2-amino-2-(hydroxymethyl)-1,3-propanediol hydrochloride (Tris HCl) and 0.1 mmol/L ethylenediaminetetraacetic acid disodium salt (disodium EDTA) in deionized water adjusted to pH 8.0 (TE -4). The three component genomic DNA materials, labeled A, B, and C, are respectively derived from a single male donor, multiple female donors, and multiple male and female donors. A unit of the SRM consists of one sterile 2-mL vial of each component, each vial containing 110 µL of DNA solution.
The SRM 2372 materials were originally certified for decadic attenuance, D10, at 230 nm, 260 nm, 270 nm, 280 nm, and 340 nm based on measurements made with a high-accuracy reference spectrophotometer. Decadic attenuance is closely related to what is commonly termed absorbance, but encompasses an instrument-dependent signal arising from optical scattering. D10 is defined from the negative base-10 logarithm of optical transmittance. The three components of the SRM were prepared to have a D10 at 260 nm very close to 1.0. Using the conventional conversion factor of 50 for double-stranded DNA (dsDNA) [Sambrook, J., Russell D.W. (2001) Molecular Cloning a Laboratory Manual, Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York], the DNA concentrations, [DNA], of the three components were about 50 ng/µL. Complete details of the production and original certification of SRM 2372 are provided elsewhere [Kline, M.C., Duewer, D.L., Travis, J.C., Smith, M.V., Redman, J.W., Vallone, P.M., Decker, A.E., Butler, J.M. (2009) Production and certification of NIST Standard Reference Material 2372 Human DNA Quantitation Standard. Anal. Bioanal. Chem. 394:1183-1192].
Unfortunately, dsDNA in TE-4 slowly acquires what can be considered to be partially single-stranded (ssDNA) character. This change in tertiary structure changes the absorbance characteristics of the solutions: ssDNA more strongly absorbs ultraviolet (UV) light than does dsDNA [Voet, D., Gratzer, W.B., Cox, R.A., Doty, P. (1963) Absorption Spectra of Nucleotides, Polynucleotides, and Nucleic Acids in the Far Ultraviolet. Bioploymers 1(3):193-208]. Thus the increase in the ssDNA character of the materials was accompanied with an increase in UV absorbance of the materials. In March 2012, SRM 2372 was restricted from sales after stability measurements of the material indicated the D10 of all three components had increased to the extent that they no longer delivered the certified D10 values within their expanded uncertainties.
We have therefore recertified the three SRM 2372 components using a method whereby high-mass DNA is forced into a pure single-stranded conformation by addition of sodium hydroxide (NaOH) [EU-JRC, Protocol NK603 – Community Reference Laboratory, Event-specific method for the quantification of maize line NK603 using real-time PCR, Section 3.3 Spectrophotometric Measurement of DNA Concentration http://gmo-crl.jrc.ec.europa.eu/summaries/NK603-WEB-ProtocolValidation.pdf]. The re-certified property for each of the SRM 2372 components is the difference in the D10 of equal-volume mixtures of the component and 0.40 mol/L aqueous NaOH at 260 nm minus the D10 at 320 nm. Subtraction of the D10 at 320 nm is an approximate correction for optical scattering. We term this property “apparent absorbance” and symbolize it “A260‑320”. Based upon A260‑320 measurements and using the conventional conversion of 37 factor for ssDNA, the [DNA] of the three components are about 60 ng/µL. This apparent increase in [DNA] suggests that when originally certified, the DNA in the SRM 2372 solutions had already acquired partial ssDNA character.
Figure 1 displays the average apparent absorbance spectra, D10 at 220 nm to 320 nm minus D10 at 320 nm, for the three SRM 2372 components. The dashed lines are the spectra for the materials as delivered in the vials, referenced to TE-4. The solid lines are the spectra for the materials diluted with an equal volume of 0.4 mol/L NaOH, referenced to TE-4 diluted with an equal volume of 0.4 mol/L NaOH.
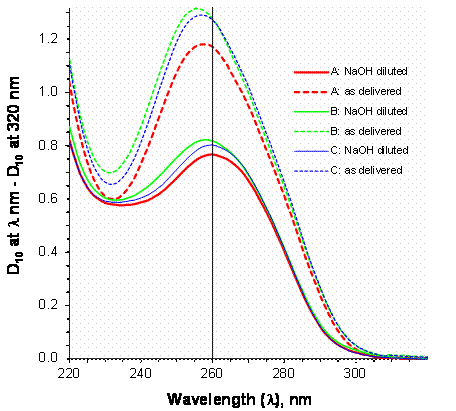
Figure 1. Apparent Absorbance
Please refer to the Certificate of Analysis for the certified values. Users of SRM 2372 should ensure that the Certificate of Analysis in their possession is current. This can be accomplished by contacting the SRM Program: telephone (301) 975 2200; fax (301) 948 3730; e mail [email protected]; or via the Internet at http://www.nist.gov/srm.
Quantitative polymerase chain reaction (qPCR) assays
The increase in ssDNA character does not affect the behavior of the materials in any quantitative polymerase chain reaction (qPCR) assay, only the inference of the conventional DNA concentration. For qPCR assays the materials should be used as delivered and NOT diluted NaOH solution.
Seven commercially available or published qPCR methods were used to analyze the relative performance of the SRM 2372 components:
Alu SYBR Green assay
Investigator Quantiplex HYres Kit (QIAGEN Inc.)
Investigator Quantiplex Kit (QIAGEN Inc.)
Plexor HY System (Promega Corporation)
Quantifiler Duo DNA Quantification Kit (Life Technologies)
Quantifiler Human DNA Quantification Kit (Life Technologies)
Quantifiler Y Human Male DNA Quantification Kit (Life Technologies)
SRM 2372 was characterized with the published Alu and CFS assays and the commercial Quantifiler Human and Quantifiler Y Human Male assays in 2006. The other assays listed above were not available at that time. The CFS assay is no longer used. Details of the experimental procedures and graphical summaries of the results are provided below.
2007 qPCR assays used a 7900HT Fast Real-Time PCR System (Applied Biosystems). The three SRM 2372 components were initially diluted 1:10 with TE-4 pH 8.0 buffer followed by serial dilutions of 1:5, 1:2, and 1:2 for a total of 4 dilutions. The Alu, CFS, Quantifiler Human, Quantifiler Y assays were 20 µL total volume. The cycling parameters for the Alu assay were: 50 °C for 2 min followed by 95 °C for 10 min followed by 30 cycles of 95 °C for 15 s, 68 °C for 30 s, 72 °C for 30 s. The CFS assay cycling parameters were: 50 °C for 2 min followed by 95 °C for 2 min, followed by 40 cycles of 95 °C for15 s, 64 °C for 30 s, 72 °C for 30 s. The cycling parameters for Quantifiler Human, and Quantifiler Y were manufacturer’s specifications.
The 2012 qPCR assays used a 7500 Real-Time PCR Instrument (Applied Biosystems). Three dilutions of components A, B, and C were prepared in TE-4 buffer at 1:10, 1:20 and 1:50 dilutions with enough volume (100 µL total volume) to use with all qPCR assays. All dilutions were run across all assays in triplicate. A no template control (NTC) was run in duplicate across all plates, and a positive control of the 1:10 dilution of component A was run in quadruplet across all plates for normalization purposes. Each dilution point (1:10, 1:20, 1:50) for each sample (A, B, C) was carried out on individual plates/runs.
The Quantifiler Human, Quantifiler Y, and Quantifiler Duo qPCR conditions included a total reaction volume of 20 µL containing 8.2 µL of the Quantifiler Human/Quantifiler Y/Quantifiler Duo Human Primer Mix and 9.8 µL of the Quantifiler Human/Quantifiler Y/Quantifiler Duo Human PCR Reaction Mix and 2 µL of the dilution of SRM 2372 component. Amplification was according to the manufacturer's recommended program: 95 ºC for 10 min, followed by 40 cycles of 15 s at 95 ºC and 1 min at 60 ºC.
The Plexor HY System conditions included a total reaction volume of 20 µL containing 10 µL Plexor HY 2X Master Mix, 1 µL Plexor HY 20X Primer/IPC Mix, 7 µL water, and 2 µL of the dilution of SRM 2372 component. Amplification was according to the manufacturer's recommended program: 95 ºC for 2 min, followed by 38 cycles of 5 s at 95 ºC and 35 s at 60 ºC, followed by a dissociation stage which consists of one cycle at 15 s at 95 ºC, 1 min at 60 ºC and 15 s at 95 ºC.
The Investigator Quantiplex qPCR conditions included a total reaction volume of 23 µL containing 11.5 µL of the Reaction Mix FQ and 11.5 µL of the Primer Mix IC FQ and 2 µL of the dilution of SRM 2372 component. Amplification was according to the manufacturer's recommended program: 95 ºC for 2 min, followed by 38 cycles of 5 s at 95 ºC and 35 s at 60 ºC.
The Investigator Quantiplex HYres qPCR included a total reaction volume of 20 µL containing 9.0 µL of the Reaction Mix FQ and 9.0 µL of the Primer Mix IC YQ and 2 µL of the dilution of SRM 2372 component. Amplification was according to the manufacturer's recommended program: 95 ºC for 3 min, followed by 40 cycles of 5 s at 95 ºC and 35 s at 60 ºC.
The in-house qPCR assay conditions included a total reaction volume of 20 µL containing 10 µL Power SYBR® Green PCR Master Mix, 0.4 µL both forward and reverse ALU primers, 7.2 µL water, and 2 µL of the dilution of SRM 2372 component. Amplification was according to the program recommended in [Nicklas, J.A., & Buel, E. (2003) Development of an Alu-based, Real-Time PCR Method for Quantitation of Human DNA in Forensic Samples. J. Forensic Sci. 48(5):936-944]: 50 ºC for 2 min, 95 ºC for 2 min, followed by 30 cycles of 15 s at 95 ºC and 30 s at 68 ºC and 45 s at 72 ºC.
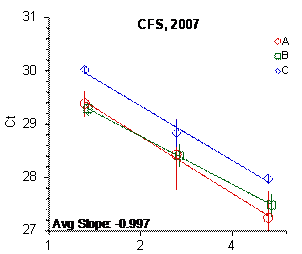
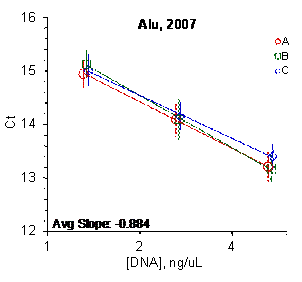
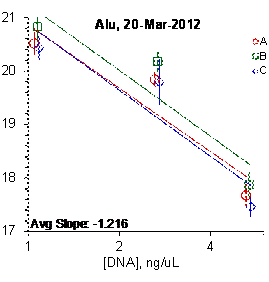
Figure 2: Results for the Alu and CFS qPCR assays
Each panel displays the average cycle threshold (Ct) as a function of DNA concentration for three dilutions of the three SRM 2372 components. The DNA concentrations are assigned from the originally certified optical density of each material multiplied by the conventional conversion factor: 50 ng/μL dsDNA = 1 D10 at 260 nm. The vertical bars are approximate 95 % level of confidence repeatability imprecision intervals.
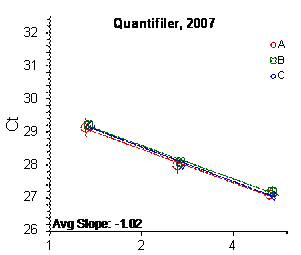
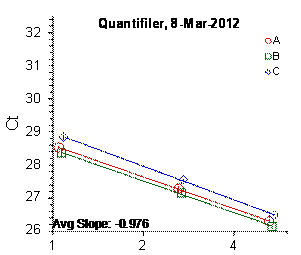
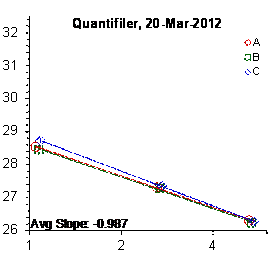
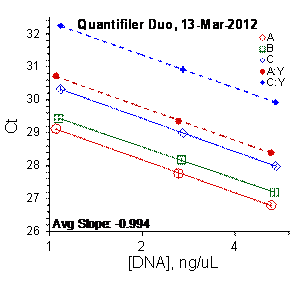
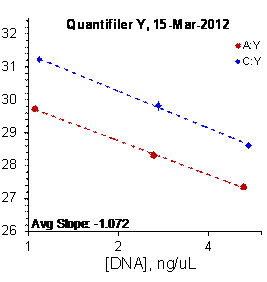
Figure 3: Results for the Quantifiler Family of qPCR Assays
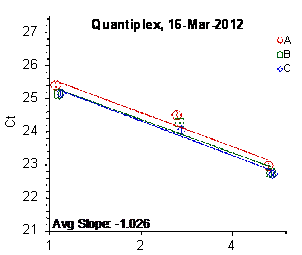
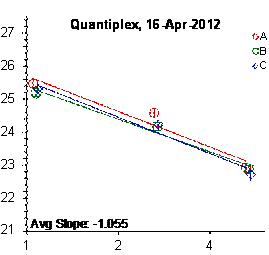
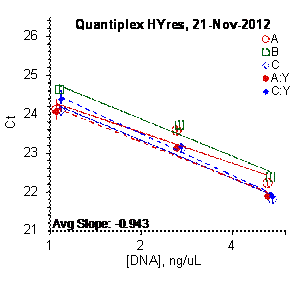
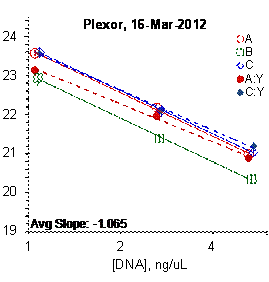
Figure 4: Results for the Quantiplex and Plexor qPCR Assays
Return to STRBase