miniSTRs (Reduced-size STR Amplicons)
NIST research in this area is being conducted by Mike Coble (at AFDIL since April 2006), Becky Hill, and John Butler with collaborator Bruce McCord (Florida International University) through funding from the National Institute of Justice.
[Introduction to miniSTRs] [Protocols and Macros for Download] [Recent Publications] [Erratum on NC01 and NC02 nomenclature]
Degraded DNA samples are commonly observed in forensic investigations involving biological evidence. Recovery of information from these degraded samples is often enhanced by use of smaller PCR products. Reduced-size STR amplicons can be created by moving the forward and reverse PCR primers in close to the STR repeat region. These so-called "miniSTR" assays can help recover information from degraded DNA samples that typically produce partial profiles and a total loss of information from larger STR amplicons. Butler, Shen, and McCord published a set of miniSTR primers that permits maximum reduction in size for all 13 CODIS STR loci along with the D2S1338, Penta D, and Penta E loci found in commercial STR kits.
A timeline for events surrounding the development and use of miniSTRs including their application in the identification of victims from the World Trade Center terrorist attacks can be found at http://www.cstl.nist.gov/biotech/strbase/miniSTR/timeline.htm. Several new miniSTR markers have been proposed to increase the number of core European STR loci--see Gill et al. (2006) The evolution of DNA databases--recommendations for new European loci. Forensic Sci. Int. 156:242-244.
Different Combinations of Loci to Create Miniplexes (published in Butler et al. (2003) J. Forensic Sci 48(5) 1054-1064.)
Using the Butler et al. (2003) published primer sequences for these miniSTR loci, a variety of combinations can be created depending on the dye label attached. A total of five miniplex combinations were described in our first paper.
| FAM | VIC | NED | ||
| "Big Mini" | miniplex 1 | TH01 | CSF1PO | TPOX |
| miniplex 3 | FGA | D21S11 | D7S820 | |
| miniplex 2 | D5S818 | D8S1179 | D16S539 | |
| miniplex 4 | VWA | D18S51 | D13S317 | |
| miniplex 5 | PentaD | PentaE | D2S1338 |
However, new combinations, such as the miniSGM assay (see below), can easily be created with the same primer sequences and the same or different dye labels. Unfortunately, due to PCR product size constraints it is not possible to usually have more than one locus per dye color. Thus, current miniplex assays are typically limited to 3 loci requiring multiple PCR amplifications to obtain equivalent numbers of loci as commercial megaplex STR assays. The use of non-nucleotide linkers to shift mobility will likely aid in creation of larger miniplexes in the future.
BodePlexes: Primer sequences and information on NIST-designed miniplexes was provided to Dr. Jim Schumm of the Bode Technology Group in April 2002 and used to create their BodePlexes (see miniSTR timeline). These BodePlexes were used starting in the fall of 2002 in aiding the World Trade Center DNA identification efforts of the New York City Office of the Chief Medical Examiner. BodePlex 1 contains D13S317, D21S11, D7S820, D16S539, and CSF1PO. BodePlex 2 contains TPOX, FGA, D7S820, and D18S51. Some limited information on the BodePlexes miniSTR assays has been published in Schumm, J.W., et al. (2004) Robust STR multiplexes for challenging casework samples. Progress in Forensic Genetics 10, ICS 1261, 547-549.
Concordance Issues (published in Drabek et al. (2004) J. Forensic Sci. 49(4): 859-860)
Due to the fact that the miniSTR primers are in different locations than the commercial STR primers, it is possible for polymorphic nucleotides or insertion/deletions in the flanking regions surrounding the STR repeats to cause allele dropout and size shifts in PCR products generated from one of the primer sets. These differences would then produce an apparent discordance. A comparison of 532 U.S. population samples (208 Caucasians, 212 African Americans, 110 Hispanics, and 2 Asians) run with both the commercial STR kit Identifiler and miniplex assays found 15 discrepancies in the 6,384 genotypes compared. These 15 discrepancies primarily involved VWA and D13S317 (see below).
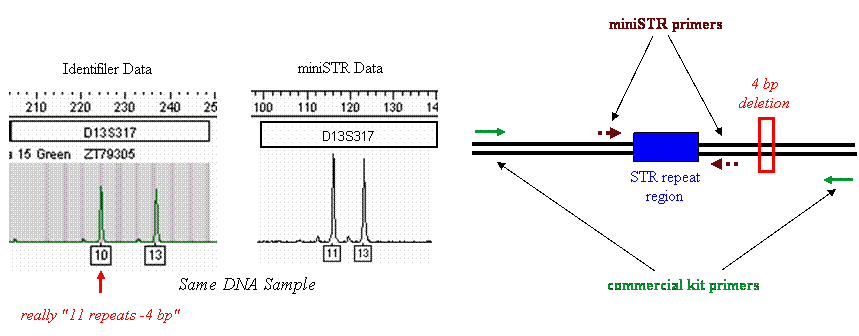
Creation of New miniSTR Loci and Assays
Since many of the commonly used CODIS STR loci contain large numbers of repeats (e.g., FGA has alleles >30 repeats in length) and are thus unable to produce PCR product sizes under 125 bp, we have begun a search for polymorphic STR loci that have smaller numbers of repeat units and fairly narrow allele ranges. Our first foray into non-CODIS (NC) loci produced a number of viable candidates and two 3plexes were produced and characterized (see Coble and Butler 2005).
More information on new NC miniSTR loci is available at http://www.cstl.nist.gov/biotech/strbase/newSTRs.htm.
Results of 26 NC miniSTR loci with standard samples, such as K562, 9947A, 9948, and 007.
Erratum on NC01 and NC02 (D22S1045, D10S1248, etc.) nomenclature:
We have changed our original D22S1045 nomenclature by +3 repeats from what was originally published in Coble and Butler (2005). This change is described in Becky Hill's poster at 17th International Symposium on Human Identification (Nashville, TN), October 10-12, 2006, "Characterization of 26 New miniSTR Loci" [.pdf]. The corrected allele range for D22S1045 should be 8-19 repeats rather than the 5-16 repeats reported previously. These changes have been also noted in Butler, J.M. and Coble, M.D. (2007) Authors' Response to Letter to Editor [regarding nomenclature for new miniSTR locus D10S1248]. J. Forensic Sci. 52(2): 494.
Reason for change: During work performed while further characterizing the NC01 loci in the spring of 2006, an error was discovered in the previously reported nomenclature for the NC01 locus D22S1045 (Coble and Butler 2005). GenBank accession G08085 contains a D22S1045 allele with 14 TAA repeats. Unfortunately, when a homozygote sample (PT84239) containing 13 TAA repeats was sequenced originally at NIST, the GenBank 14 allele and the PT84239 13 allele were accidentally inverted in the nomenclature notes. Thus, Coble and Butler (2005) reported the GenBank accession G08085 allele to be 13 TAA repeats (see Table 1 in Coble and Butler 2005--it should have been 14 TAA repeats). However, the Genotyping macros and allelic ladders provided to collaborating labs (see Figure 3 in Coble and Butler 2005) define an allele range of 5-16 repeats, which correctly reflected the actual sequences. We have decided to change to the opposite strand in using GenBank accession AL022314 as a reference sequence, which changes the repeat unit from TAA to ATT. In addition, after further review of D22S1045 and other miniSTR loci, we have decided to change our original nomenclature and add an additional 1 ACT and 2 ATT repeats. Thus, the new allelic ladder range following this adjustment will be 8-19 repeats.
The original D10S1248 nomenclature has been reduced by -1 repeat from what was originally published in Coble and Butler (2005). The corrected allele range for D10S1248 should be 9-19 repeats rather than the 10-20 repeats reported previously. These changes have been also noted in Butler, J.M. and Coble, M.D. (2007) Authors' Response to Letter to Editor [regarding nomenclature for new miniSTR locus D10S1248]. J. Forensic Sci. 52(2): 494.
Reason for change: Further sequence analysis conducted in the summer of 2006...
The original D14S1434 nomenclature has been reduced by -4 repeats from what was originally published in Coble and Butler (2005). The corrected allele range for D14S1434 should be 9-16 repeats rather than the 13-20 repeats reported previously. These changes have been also noted in Butler, J.M. and Coble, M.D. (2007) Authors' Response to Letter to Editor [regarding nomenclature for new miniSTR locus D10S1248]. J. Forensic Sci. 52(2): 494.
Reason for change: Further sequence analysis conducted in the summer and fall of 2006...
The original D1S1677 nomenclature has been increased by +1 repeat from what was originally published in Coble and Butler (2005). The corrected allele range for D1S1677 should be 10-19 repeats rather than the 9-18 repeats reported previously. These changes have been also noted in Butler, J.M. and Coble, M.D. (2007) Authors' Response to Letter to Editor [regarding nomenclature for new miniSTR locus D10S1248]. J. Forensic Sci. 52(2): 494.
Reason for change: Further sequence analysis conducted in the summer and fall of 2006...
The original D4S2364 nomenclature has been reduced by -1 repeat from what was originally published in Coble and Butler (2005). The corrected allele range for D4S2364 should be 7-11 repeats rather than the 8-12 repeats reported previously. These changes have been also noted in Butler, J.M. and Coble, M.D. (2007) Authors' Response to Letter to Editor [regarding nomenclature for new miniSTR locus D10S1248]. J. Forensic Sci. 52(2): 494.
Reason for change: Further sequence analysis conducted in the summer and fall of 2006...
Protocols and Genotyper Macros for Download
| Assay Name | miniSGM | miniNC01 |
| Loci Amplified | TH01, Amelogenin, FGA, D18S51, D16S539, D2S11338 | D10S1248, D14S1434, D22S1045 |
| Protocol | [click to download] | [updated protocol] |
| Genotyper NT Macro | [click to download] | [click to download] |
| Allele Sizes | [click to download] | [click to download] |
Protocol for creation of miniSTR allelic ladders from existing allelic ladders
![]() GeneMapperID
v3.2 Bins and Panels for 26 NC miniSTR Loci (used with ABI 3100 POP6): [panels]
[bins]
GeneMapperID
v3.2 Bins and Panels for 26 NC miniSTR Loci (used with ABI 3100 POP6): [panels]
[bins]
Bruce McCord Laboratory: [Protocols for Big Mini, Mini2, and Mini4] [Miniplex STR Amplification Worksheet]
| Assay Name | Big Mini | Mini2 | Mini4 |
| Loci Amplified | TH01, FGA, CSF1PO, D21S11, TPOX, D7S820 | D5S818, D8S1179, D16S539 | vWA, D18S51, D13S317 |
| Protocol | [click to download] (contact Bruce McCord <[email protected]> for more details) | ||
| Genotyper NT Macro | [click to download] | [click to download] | [click to download] |
![]() GeneMapperID
v3.2 Bins and Panels for 12 CODIS miniSTR Loci in BigMini, Mini2, and Mini4: [panels]
[bins]
GeneMapperID
v3.2 Bins and Panels for 12 CODIS miniSTR Loci in BigMini, Mini2, and Mini4: [panels]
[bins]
For more information, contact: John Butler, NIST-Biochemical Science Division, 301-975-4049, [email protected]
Recent Publications on miniSTRs
Butler, J.M., Shen, Y., McCord, B.R. (2003) The development of reduced size STR amplicons as tools for analysis of degraded DNA. J. Forensic Sci 48(5) 1054-1064.
Chung, D.T., Drabek, J., Opel, K.L., Butler, J.M., McCord, B.R. (2004) A study on the effects of degradation and template concentration on the efficiency of the STR miniplex primer sets. J. Forensic Sci. 49(4): 733-740.
Drabek, J., Chung, D.T., Butler, J.M., McCord, B.R. (2004) Concordance study between miniplex STR assays and a commercial STR typing kit, J. Forensic Sci. 49(4): 859-860.
Coble, M.D. and Butler, J.M. (2005) Characterization of new miniSTR loci to aid analysis of degraded DNA. J. Forensic Sci. 50(1):43-53.
Coble, M.D., Hill, C.R., Vallone, P.M., Butler, J.M. (2006) Characterization and performance of new miniSTR loci for typing degraded samples. Progress in Forensic Genetics 11, Elsevier Science: Amsterdam, The Netherlands, International Congress Series 1288, 504-506.
Becky Hill poster at 58th Annual Meeting of the American Academy of Forensic Sciences (Seattle, WA), February 24, 2006, "Development of 27 New miniSTR Loci for Improved Analysis of Degraded DNA Samples" [.pdf]
Opel, K.L., Chung, D.T., Drabek, J., Tatarek, N.E., Jantz, L.M., McCord, B.R. (2006) The application of miniplex primer sets in the analysis of degraded DNA from human skeletal remains. J. Forensic Sci. 51(2): 351-356.
Hill, C.R., Kline, M.C., Coble, M.D., Butler, J.M. (2006) Characterization of 26 miniSTR loci for improved analysis of degraded DNA samples. submitted.
Becky Hill poster at 17th International Symposium on Human Identification (Nashville, TN), October 10-12, 2006, "Characterization of 26 New miniSTR Loci" [.pdf]
Margaret Kline poster at 17th International Symposium on Human Identification (Nashville, TN), October 10-12, 2006, "NIST SRM Updates: Value-added to the Current Materials in SRM 2391b and SRM 2395" [.pdf]
Butler, J.M. (2006) MiniSTRs: past, present, and future. Forensic News (Applied Biosystems), October 2006 [.pdf]
Dixon, L.A., Dobbins, A.E., Pulker, H., Butler, J.M., Vallone, P.M., Coble, M.D., Parson, W., Berger, B., Grubweiser, P., Mogensen, H.S., Morling, N., Nielsen, K., Sanchez, J.J., Petkovski, E., Carracedo, A., Sanchez-Diz, P., Brion, M., Irwin, J.A., Just, R.S., Loreille, O., Parsons, T.J., Syndercombe-Court, D., Schmitter, H., Gill, P. (2006) Analysis of artificially degraded DNA using STRs and SNPs--results of a collaborative European (EDNAP) exercise. Forensic Sci. Int. 164: 33-44.
Yong, R.Y.Y., Gan, L.S.H., Coble, M.D., Yap, E.P.H. (2007) Allele frequencies of six miniSTR loci of three ethnic populations in Singapore. Forensic Sci. Int. 166: 240-243.
Parsons, T.J., Huel, R., Davoren, J., Katzmarzyk, C., Milos, A., Selmanovic, A., Smajlovic, L., Coble, M.D., Rizvic, A. (2007) Application of novel "mini-amplicon" STR multiplexes to high volume casework on degraded skeletal remains. FSI Genetics 1:175-179.
Hill, C.R., Kline, M.C., Mulero, J.J., Lagace, R.E., Chang, C.-W., Hennessy, L.K., Butler, J.M. (2007) Concordance study between the AmpFlSTR MiniFiler PCR Amplification Kit and conventional STR typing kits. J. Forensic Sci. 52(4): 870-873.
Work by others on miniSTRs:
Gill, P., Fereday, L., Morling, N., Schneider, P.M. (2006) The evolution of DNA databases--recommendations for new European loci. Forensic Sci. Int. 156:242-244.
Gill, P., Fereday, L., Morling, N., Schneider, P.M. (2006) Letter to editor -- New multiplexes for Europe-amendments and clarification of strategic development. Forensic Sci Int. 163:155-157.
Grubwieser, P., Muhlmann, R., and Parson, W. (2003) New sensitive amplification primers for the STR locus D2S1338 for degraded casework DNA. Int. J. Legal Med. 117(3): 185-188.
Grubwieser, P., Muhlmann, R., Berger, B., Niederstatter, H., Pavlic, M., Parson, W. (2006) A new "miniSTR-multiplex" displaying reduced amplicon lengths for the analysis of degraded DNA. Int. J. Legal Med. 120:115-120.
Hellmann, A., Rohleder, U., Schmitter, H., and Wittig, M. (2001) STR typing of human telogen hairs--a new approach. Int. J. Legal Med. 114(4-5): 269-273.
Holland, M.M., Cave, C.A., Holland, C.A. and Bille, T.W. Development of a quality, high throughput DNA analysis procedure for skeletal samples to assist with the identification of victims from the World Trade Center attacks.(2003) Croatian Medical Journal 44:264-272.
Ohtaki, H., Yamamoto, T., Yoshimoto, T., Uchihi, R., Ooshima, C., Katsumata, Y., and Tokunaga, K. (2002) A powerful, novel, multiplex typing system for six short tandem repeat loci and the allele frequency distributions in two Japanese regional populations. Electrophoresis 23(19): 3332-3340.
Schumm, J.W., Wingrove, R.S., Douglas, E.K. (2004) Robust STR multiplexes for challenging casework samples. Progress in Forensic Genetics ICS 1261:547-549
Szibor, R., Kayser, M., and Roewer, L. (2000) Identification of the human Y-chromosomal microsatellite locus DYS19 from degraded DNA. Am. J .Forensic Med. Pathol. 21(3): 252-254.
Tsukada, K., Takayanagi, K., Asamura, H., Ota, M., and Fukushima, H. (2002) Multiplex short tandem repeat typing in degraded samples using newly designed primers for the TH01,CSF1PO,and vWA loci. Legal Medicine 4: 239-245.
Wiegand, P. and Kleiber, M. (2001) Less is more--length reduction of STR amplicons using redesigned primers. Int. J. Legal Med. 114(4-5): 285-287.
Wiegand P, Klein R, Braunschweiger G, Hohoff C, Brinkmann B. (2006) Short amplicon STR multiplex for stain typing. Int. J. Legal Med. 120(3):160-164
Yoshida, K., Sekiguchi, K., Kasai, K., Sato, H., Seta, S., and Sensabaugh, G. F. (1997) Evaluation of new primers for CSF1PO. Int. J. Legal Med. 110(1): 36-38.
Applied Biosystems MiniFiler kit
October 2006 Forensic News: http://marketing.appliedbiosystems.com/images/enews/ForensicNews_Vol7/PDF/00_ForensicNews.pdf
MiniFiler product information: http://minifiler.appliedbiosystems.com
Hill, C.R., Kline, M.C., Mulero, J.J., Lagace, R.E., Chang, C.-W., Hennessy, L.K., Butler, J.M. (2007) Concordance study between the AmpFlSTR MiniFiler PCR Amplification Kit and conventional STR typing kits. J. Forensic Sci. 52(4): 870-873.
For other NIST human identity project team publications, see http://www.cstl.nist.gov/biotech/strbase/NISTpub.htm.