SRM 2391b PCR-based DNA Profiling Standard
NOW AVAILABLE: see http://www.nist.gov/srm
https://www-s.nist.gov/srmors/view_detail.cfm?srm=2391B
The Certificate of Analysis for SRM 2391b has been updated to include information from 26 new STR loci as well as the MiniFiler kit.
For further information on the new STR loci, see Hill, C.R., Kline, M.C., Coble, M.D., Butler, J.M. (2008) Characterization of 26 miniSTR loci for improved analysis of degraded DNA samples. J. Forensic Sci. 53(1):73-80.
[Supporting
Sequence Information] [Stability Information]
Supporting Sequence Information
Allele assignments with associated repeat motifs established by sequencing are considered Certified Values. Those allele assignments without sequence established repeat motifs are considered Reference Values.
|
|
|
D4S2364 |
|
|
|
D10S1248 |
|
|
|
D14S1434 |
|
|
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
1 |
9 |
[ATTC]7 ATCC ATTC |
75.33 |
|
14 |
[GGAA]14 |
103.51 |
|
13 |
[CTGT]3 [CTAT]10 |
86.28 |
|
|
|
|
|
|
16 |
[GGAA]16 |
111.34 |
|
14 |
[CTGT]3 [CTAT]11 |
90.49 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2 |
9 |
[ATTC]7 ATCC ATTC |
75.33 |
|
13 |
[GGAA]13 |
99.52 |
|
11 |
[CTGT]3 [CTAT]8 |
77.96 |
|
|
10 |
[ATTC]8 ATCC ATTC |
79.38 |
|
15 |
[GGAA]15 |
107.38 |
|
13 |
[CTGT]3 [CTAT]10 |
86.23 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3 |
9 |
[ATTC]7 ATCC ATTC |
75.33 |
|
13 |
[GGAA]13 |
99.52 |
|
14 |
[CTGT]3 [CTAT]11 |
90.42 |
|
|
10 |
[ATTC]8 ATCC ATTC |
79.41 |
|
16 |
[GGAA]16 |
111.30 |
|
15 |
[CTGT]3 [CTAT]12 |
94.64 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
4 |
9 |
[ATTC]7 ATCC ATTC |
75.34 |
|
12 |
[GGAA]12 |
95.31 |
|
10 |
[CTGT]3 [CTAT]7 |
73.84 |
|
|
|
|
|
|
|
|
|
|
11 |
[CTGT]3 [CTAT]8 |
77.87 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5 |
9 |
[ATTC]7 ATCC ATTC |
75.30 |
|
14 |
[GGAA]14 |
103.48 |
|
13 |
[CTGT]3 [CTAT]10 |
86.32 |
|
|
10 |
[ATTC]8 ATCC ATTC |
79.37 |
|
15 |
[GGAA]15 |
107.40 |
|
14 |
[CTGT]3 [CTAT]11 |
90.51 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
6 |
8 |
[ATTC]6 ATCC ATTC |
71.38 |
|
14 |
[GGAA]14 |
103.50 |
|
13 |
[CTGT]3 [CTAT]10 |
86.29 |
|
|
9 |
[ATTC]7 ATCC ATTC |
75.30 |
|
15 |
[GGAA]15 |
107.38 |
|
14 |
[CTGT]3 [CTAT]11 |
90.46 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
7 |
9 |
[ATTC]7 ATCC ATTC |
75.26 |
|
13 |
[GGAA]13 |
99.56 |
|
10 |
[CTGT]3 [CTAT]7 |
73.90 |
|
|
|
|
|
|
14 |
[GGAA]14 |
103.51 |
|
14 |
[CTGT]3 [CTAT]11 |
90.45 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
8 |
9 |
[ATTC]7 ATCC ATTC |
75.35 |
|
11 |
[GGAA]11 |
91.17 |
|
13 |
[CTGT]3 [CTAT]10 |
86.25 |
|
|
|
|
|
|
15 |
[GGAA]15 |
107.38 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
9 |
9 |
[ATTC]7 ATCC ATTC |
75.37 |
|
13 |
[GGAA]13 |
99.58 |
|
11 |
[CTGT]3 [CTAT]8 |
78.04 |
|
|
10 |
[ATTC]8 ATCC ATTC |
79.43 |
|
15 |
[GGAA]15 |
107.43 |
|
13 |
[CTGT]3 [CTAT]10 |
86.31 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
10 |
9 |
[ATTC]7 ATCC ATTC |
75.33 |
|
12 |
[GGAA]12 |
95.37 |
|
13 |
[CTGT]3 [CTAT]10 |
86.33 |
|
|
10 |
[ATTC]8 ATCC ATTC |
79.38 |
|
15 |
[GGAA]15 |
107.44 |
|
14 |
[CTGT]3 [CTAT]11 |
90.51 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
11 |
9 |
|
74.94* |
|
13 |
|
99.32* |
|
11 |
|
77.49* |
|
|
10 |
|
78.99* |
|
15 |
|
107.18* |
|
13 |
|
85.68* |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
12 |
9 |
|
74.94* |
|
12 |
|
95.09* |
|
13 |
|
85.63* |
|
|
10 |
|
78.99* |
|
15 |
|
107.19* |
|
14 |
|
89.81* |
|
|
|
D1S1677 |
|
|
|
D2S441 |
|
|
|
D22S1045 |
|
|
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
1 |
12 |
[TTCC]12 |
89.66 |
|
11 |
[TCTA]11 |
86.04 |
|
14 |
[ATT]11 ACT [ATT]2 |
94.45 |
|
|
13 |
|
93.79 |
|
14 |
[TCTA]11 TTTA [TCTA]2 |
98.69 |
|
15 |
[ATT]12 ACT [ATT]2 |
97.63 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2 |
14 |
[TTCC]14 |
97.99 |
|
11 |
[TCTA]11 |
86.05 |
|
11 |
[ATT]8 ACT [ATT]2 |
84.89 |
|
|
16 |
[TTCC]16 |
105.97 |
|
14 |
[TCTA]11 TTTA [TCTA]2 |
98.69 |
|
16 |
|
100.68 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3 |
14 |
[TTCC]14 |
97.99 |
|
10 |
[TCTA]8 TCTG TCTA |
82.05 |
|
15 |
[ATT]12 ACT [ATT]2 |
97.55 |
|
|
17 |
[TTCC]17 |
109.91 |
|
14 |
[TCTA]11 TTTA [TCTA]2 |
98.73 |
|
16 |
|
100.69 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
4 |
14 |
[TTCC]14 |
97.96 |
|
12 |
|
90.20 |
|
17 |
|
103.62 |
|
|
15 |
[TTCC]15 |
102.01 |
|
14 |
[TCTA]11 TTTA [TCTA]2 |
98.67 |
|
18 |
[ATT]15 ACT [ATT]2 |
106.61 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5 |
13 |
[TTCC]13 |
93.71 |
|
11 |
[TCTA]11 |
85.98 |
|
11 |
[ATT]8 ACT [ATT]2 |
85.04 |
|
|
14 |
[TTCC]14 |
97.88 |
|
14 |
[TCTA]11 TTTA [TCTA]2 |
98.60 |
|
14 |
[ATT]11 ACT [ATT]2 |
94.46 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
6 |
13 |
[TTCC]13 |
93.67 |
|
10 |
[TCTA]10 |
81.85 |
|
11 |
[ATT]8 ACT [ATT]2 |
84.98 |
|
|
14 |
[TTCC]14 |
97.86 |
|
11 |
[TCTA]11 |
85.97 |
|
15 |
[ATT]12 ACT [ATT]2 |
97.60 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
7 |
12 |
[TTCC]12 |
89.53 |
|
11 |
[TCTA]11 |
85.94 |
|
11 |
[ATT]8 ACT [ATT]2 |
84.95 |
|
|
13 |
|
93.70 |
|
14 |
[TCTA]11 TTTA [TCTA]2 |
98.59 |
|
15 |
[ATT]12 ACT [ATT]2 |
97.62 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
8 |
14 |
[TTCC]14 |
98.00 |
|
11 |
|
86.09 |
|
16 |
[ATT]13 ACT [ATT]2 |
100.75 |
|
|
16 |
[TTCC]16 |
105.97 |
|
11.3 |
[TCTA]4 TCA [TCTA]7 |
89.14 |
|
17 |
[ATT]14 ACT [ATT]2 |
103.72 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
9 |
13 |
[TTCC]13 |
93.85 |
|
10 |
[TCTA]8 TCTG TCTA |
82.12 |
|
11 |
[ATT]8 ACT [ATT]2 |
84.97 |
|
|
14 |
[TTCC]14 |
98.03 |
|
14 |
[TCTA]11 TTTA [TCTA]2 |
98.76 |
|
14 |
[ATT]11 ACT [ATT]2 |
94.47 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
10 |
13 |
[TTCC]13 |
93.80 |
|
11 |
[TCTA]11 |
86.06 |
|
16 |
|
100.81 |
|
|
14 |
[TTCC]14 |
98.02 |
|
12 |
[TCTA]12 |
90.12 |
|
18 |
[ATT]15 ACT [ATT]2 |
106.75 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
11 |
13 |
|
93.22* |
|
10 |
|
81.61* |
|
11 |
|
84.42* |
|
|
14 |
|
97.39* |
|
14 |
|
98.13* |
|
14 |
|
93.82* |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
12 |
13 |
|
93.24* |
|
11 |
|
85.56* |
|
16 |
|
100.07* |
|
|
14 |
|
97.46* |
|
12 |
|
89.74* |
|
18 |
|
106.05* |
|
|
|
D1GATA113 |
|
|
|
D1S1627 |
|
|
|
D2S1776 |
|
|
|
D3S3053 |
|
|
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
1 |
11 |
[GATA]11 |
97.57 |
|
10 |
[ATT]10 |
81.16 |
|
11 |
|
144.33 |
|
9 |
|
92.89 |
|
|
|
|
|
|
14 |
[ATT]14 |
93.68 |
|
12 |
|
148.79 |
|
12 |
|
105.18 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2 |
12 |
|
101.66 |
|
13 |
|
90.56 |
|
11 |
[AGAT]11 |
144.34 |
|
10 |
|
97.32 |
|
|
13 |
|
105.58 |
|
14 |
|
93.71 |
|
|
|
|
|
11 |
|
101.48 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3 |
11 |
[GATA]11 |
97.51 |
|
13 |
|
90.52 |
|
8 |
|
131.54 |
|
9 |
|
92.93 |
|
|
|
|
|
|
14 |
|
93.70 |
|
10 |
|
139.86 |
|
11 |
|
101.30 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
4 |
13 |
|
105.55 |
|
11 |
|
84.27 |
|
11 |
|
144.34 |
|
11 |
[TATC]11 |
101.31 |
|
|
|
|
|
|
12 |
|
87.39 |
|
12 |
|
148.75 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5 |
11 |
|
97.56 |
|
14 |
|
93.69 |
|
12 |
|
148.76 |
|
11 |
[TATC]11 |
101.31 |
|
|
12 |
|
101.64 |
|
15 |
|
96.83 |
|
13 |
|
153.09 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
6 |
11 |
|
97.58 |
|
11 |
|
84.22 |
|
11 |
|
144.37 |
|
9 |
[TATC]9 |
92.94 |
|
|
12 |
|
101.71 |
|
13 |
|
90.50 |
|
12 |
|
148.81 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
7 |
10 |
|
93.37 |
|
11 |
[ATT]11 |
84.23 |
|
11 |
|
144.35 |
|
11 |
[TATC]11 |
101.29 |
|
|
12 |
|
101.68 |
|
14 |
[ATT]14 |
93.66 |
|
12 |
|
148.76 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
8 |
10 |
|
93.37 |
|
13 |
|
90.51 |
|
11 |
|
144.37 |
|
9 |
[TATC]9 |
92.87 |
|
|
12 |
|
101.68 |
|
14 |
|
93.68 |
|
12 |
|
148.80 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
9 |
11 |
|
97.51 |
|
13 |
[ATT]13 |
90.54 |
|
10 |
[AGAT]10 |
139.88 |
|
9 |
|
92.96 |
|
|
12 |
|
101.63 |
|
14 |
[ATT]14 |
93.71 |
|
|
|
|
|
11 |
|
101.32 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
10 |
7 |
|
80.92 |
|
11 |
[ATT]11 |
84.24 |
|
10 |
|
139.90 |
|
9 |
|
92.91 |
|
|
12 |
|
101.73 |
|
13 |
[ATT]13 |
90.50 |
|
12 |
|
148.81 |
|
12 |
|
105.22 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
11 |
11 |
|
97.06* |
|
13 |
|
89.80* |
|
10 |
[AGAT]10 |
139.55* |
|
9 |
|
92.40* |
|
|
12 |
|
101.20* |
|
14 |
|
92.91* |
|
|
|
|
|
11 |
|
100.80* |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
12 |
7 |
|
80.47* |
|
11 |
|
83.59* |
|
10 |
|
139.64* |
|
9 |
|
92.41* |
|
|
12 |
|
101.21* |
|
13 |
|
89.80* |
|
12 |
|
148.50* |
|
12 |
|
104.73* |
|
|
|
D3S4529 |
|
|
|
D4S2408 |
|
|
|
D5S2500 |
|
|
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
1 |
14 |
|
123.24 |
|
10 |
[ATCT]10 |
97.31 |
|
17 |
|
97.47 |
|
|
15 |
|
127.37 |
|
|
|
|
|
18 |
|
101.47 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2 |
13 |
|
199.17 |
|
9 |
[ATCT]9 |
93.07 |
|
17 |
[GGTA]3 [GACA]8 [GATA]3 [GATT]3 |
97.46 |
|
|
16 |
|
131.42 |
|
|
|
|
|
24 |
[GGTA]10 [GACA]6 [GATA]3 [GATT]5 |
126.29 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3 |
14 |
|
123.24 |
|
8 |
|
88.87 |
|
17 |
|
97.44 |
|
|
16 |
|
131.45 |
|
9 |
|
93.05 |
|
18 |
|
101.47 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
4 |
15 |
|
127.45 |
|
9 |
|
93.05 |
|
17 |
|
97.41 |
|
|
16 |
|
131.56 |
|
10 |
|
97.23 |
|
18 |
|
101.41 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5 |
13 |
|
119.29 |
|
10 |
|
97.29 |
|
14 |
|
85.04 |
|
|
15 |
|
127.46 |
|
11 |
|
101.43 |
|
15 |
|
89.09 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
6 |
15 |
|
127.4 |
|
9 |
[ATCT]9 |
93.12 |
|
14 |
[GGTA]3 [GACA]6 [GATA]2 [GATT]3 |
85.06 |
|
|
17 |
|
135.66 |
|
|
|
|
|
18 |
[GGTA]3 [GACA]9 [GATA]3 [GATT]3 |
101.47 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
7 |
14 |
|
123.32 |
|
8 |
|
88.95 |
|
14 |
[GGTA]3 [GACA]6 [GATA]2 [GATT]3 |
85.04 |
|
|
16 |
|
131.49 |
|
11 |
|
101.47 |
|
20 |
[GGTA]8 [GACA]5 [GATA]3 [GATT]4 |
110.15 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
8 |
14 |
[ATCT]9 ATTT [ATCT]4 |
123.32 |
|
11 |
[ATCT]11 |
101.47 |
|
14 |
|
85.08 |
|
|
|
|
|
|
|
|
|
|
18 |
|
101.46 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
9 |
13 |
[ATCT]8 ATTT [ATCT]4 |
119.31 |
|
9 |
|
93.04 |
|
14 |
[GGTA]3 [GACA]6 [GATA]2 [GATT]3 |
85.06 |
|
|
|
|
|
|
10 |
|
97.24 |
|
23 |
[GGTA]9 [GACA]6 [GATA]3 [GATT]5 |
121.14 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
10 |
12 |
[ATCT]7 ATTT [ATCT]4 |
115.24 |
|
10 |
[ATCT]10 |
97.43 |
|
14 |
[GGTA]4 [GACA]6 [GATA]1 [GATT]3 |
85.35 |
|
|
|
|
|
|
|
|
|
|
17 |
[GGTA]4 [GACA]7 [GATA]3 [GATT]3 |
97.40 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
11 |
13 |
[ATCT]8 ATTT [ATCT]4 |
118.47* |
|
9 |
|
92.56* |
|
14 |
|
84.69* |
|
|
|
|
|
|
10 |
|
96.76* |
|
23 |
|
121.85* |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
12 |
12 |
[ATCT]7 ATTT [ATCT]4 |
114.51* |
|
10 |
[ATCT]10 |
96.72* |
|
14 |
|
84.72* |
|
|
|
|
|
|
|
|
|
|
17 |
|
97.05* |
|
|
|
D6S474 |
|
|
|
D6S1017 |
|
|
|
D8S1115 |
|
|
|
D9S1122 |
|
|
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
1 |
15 |
|
116.12 |
|
10 |
[ATCC]10 |
93.66 |
|
16 |
[ATT]16 |
83.32 |
|
11 |
|
101.27 |
|
|
17 |
|
124.17 |
|
|
|
|
|
|
|
|
|
12 |
|
105.21 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2 |
14 |
|
112.26 |
|
10 |
|
93.64 |
|
16 |
[ATT]16 |
83.29 |
|
12 |
|
105.27 |
|
|
17 |
|
124.35 |
|
12 |
|
101.80 |
|
|
|
|
|
13 |
|
109.22 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3 |
14 |
|
112.23 |
|
10 |
|
93.61 |
|
16 |
|
83.30 |
|
12 |
TAGA TCGA [TAGA]10 |
105.24 |
|
|
15 |
|
116.21 |
|
12 |
|
101.80 |
|
17 |
|
86.44 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
4 |
14 |
|
112.21 |
|
7 |
|
81.35 |
|
9 |
|
62.34 |
|
12 |
TAGA TCGA [TAGA]10 |
105.20 |
|
|
16 |
|
120.19 |
|
10 |
|
93.61 |
|
17 |
|
86.46 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5 |
15 |
|
116.23 |
|
8 |
|
85.39 |
|
9 |
|
62.35 |
|
11 |
|
101.28 |
|
|
18 |
|
128.32 |
|
9 |
|
89.49 |
|
15 |
|
80.20 |
|
13 |
|
109.12 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
6 |
14 |
|
112.21 |
|
10 |
[ATCC]10 |
93.60 |
|
9 |
|
62.32 |
|
11 |
|
101.29 |
|
|
17 |
|
124.29 |
|
|
|
|
|
16 |
|
83.34 |
|
12 |
|
105.20 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
7 |
15 |
|
116.19 |
|
7 |
|
81.38 |
|
9 |
|
62.32 |
|
11 |
|
101.34 |
|
|
17 |
|
124.26 |
|
12 |
|
101.76 |
|
18 |
|
89.63 |
|
12 |
|
105.25 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
8 |
17 |
[AGAT]5 [GATA]12 |
124.20 |
|
10 |
|
93.64 |
|
15 |
|
80.23 |
|
13 |
TAGA TCGA [TAGA]11 |
109.19 |
|
|
|
|
|
|
12 |
|
101.79 |
|
16 |
|
83.32 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
9 |
14 |
|
112.21 |
|
9 |
|
89.49 |
|
9 |
|
62.33 |
|
12 |
|
105.22 |
|
|
18 |
|
128.33 |
|
10 |
|
93.61 |
|
18 |
|
89.61 |
|
13 |
|
109.20 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
10 |
17 |
[AGAT]5 [GATA]12 |
124.28 |
|
8 |
[ATCC]8 |
85.52 |
|
15 |
|
80.21 |
|
12 |
|
105.26 |
|
|
|
|
|
|
|
|
|
|
17 |
|
86.50 |
|
15 |
|
117.21 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
11 |
14 |
|
111.93* |
|
9 |
|
88.87* |
|
9 |
|
62.24* |
|
12 |
|
104.97* |
|
|
18 |
|
128.19* |
|
10 |
|
92.91* |
|
18 |
|
88.92* |
|
13 |
|
108.91* |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
12 |
17 |
[AGAT]5 [GATA]12 |
124.06* |
|
8 |
[ATCC]8 |
84.86* |
|
15 |
|
79.58* |
|
12 |
|
104.97* |
|
|
|
|
|
|
|
|
|
|
17 |
|
85.83* |
|
15 |
|
116.99* |
|
|
|
D9S2157 |
|
|
|
D10S1435 |
|
|
|
D11S4463 |
|
|
|
D12ATA63 |
|
|
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
1 |
8 |
|
73.46 |
|
13 |
[TATC]13 |
114.83 |
|
14 |
[TATC]12 TATG TATC |
103.77 |
|
14 |
|
88.00 |
|
|
13 |
|
88.78 |
|
|
|
|
|
|
|
|
|
17 |
|
97.40 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2 |
9 |
|
76.47 |
|
11 |
|
106.83 |
|
13 |
|
99.91 |
|
13 |
|
84.89 |
|
|
11 |
|
82.60 |
|
14 |
|
118.81 |
|
14 |
|
103.75 |
|
17 |
|
97.37 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3 |
11 |
|
82.60 |
|
13 |
|
114.79 |
|
14 |
|
103.88 |
|
12 |
|
81.83 |
|
|
13 |
|
88.77 |
|
14 |
|
118.81 |
|
15 |
|
107.75 |
|
15 |
|
91.04 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
4 |
11 |
[ATA]11 |
82.65 |
|
12 |
[TATC]12 |
110.80 |
|
11 |
|
91.47 |
|
16 |
|
94.22 |
|
|
|
|
|
|
|
|
|
|
12 |
|
95.72 |
|
18 |
|
100.47 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5 |
7 |
|
70.60 |
|
11 |
|
106.75 |
|
13 |
|
99.87 |
|
13 |
|
84.89 |
|
|
14 |
|
91.93 |
|
12 |
|
110.74 |
|
15 |
|
107.79 |
|
15 |
|
91.10 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
6 |
11 |
|
82.61 |
|
12 |
[TATC]12 |
110.76 |
|
15 |
|
107.77 |
|
14 |
|
87.94 |
|
|
13 |
|
88.79 |
|
|
|
|
|
16 |
|
111.90 |
|
18 |
|
100.47 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
7 |
12 |
|
85.68 |
|
12 |
[TATC]12 |
110.79 |
|
13 |
|
99.87 |
|
16 |
|
94.29 |
|
|
15 |
|
95.08 |
|
|
|
|
|
14 |
|
103.93 |
|
17 |
|
97.43 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
8 |
11 |
[ATA]11 |
82.62 |
|
11 |
|
106.80 |
|
13 |
|
99.87 |
|
14 |
|
87.99 |
|
|
|
|
|
|
13 |
|
114.78 |
|
16 |
|
111.77 |
|
15 |
|
91.01 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
9 |
7 |
|
70.61 |
|
10 |
|
102.84 |
|
12 |
|
95.80 |
|
13 |
[TAA]10 [CAA]3 |
84.87 |
|
|
13 |
|
88.77 |
|
11 |
|
106.80 |
|
13 |
|
100.04 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
10 |
7 |
|
70.60 |
|
12 |
|
110.75 |
|
12 |
|
95.65 |
|
13 |
|
84.91 |
|
|
11 |
|
82.61 |
|
13 |
|
114.88 |
|
14 |
|
103.95 |
|
18 |
|
100.53 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
11 |
7 |
|
70.41* |
|
10 |
|
102.17* |
|
12 |
|
95.08* |
|
13 |
[TAA]10 [CAA]3 |
84.47* |
|
|
13 |
|
88.21* |
|
11 |
|
106.16* |
|
13 |
|
99.29* |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
12 |
7 |
|
70.41* |
|
12 |
|
110.21* |
|
12 |
|
94.94* |
|
13 |
|
84.53* |
|
|
11 |
|
82.10* |
|
13 |
|
114.24* |
|
14 |
|
103.31* |
|
18 |
|
100.07* |
|
|
|
D17S974 |
|
|
|
D17S1301 |
|
|
|
D18S853 |
|
|
|
D20S482 |
|
|
|
D20S1082 |
|
|
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
Type |
Repeat motif |
Size |
|
1 |
9 |
|
111.17 |
|
11 |
|
122.48 |
|
11 |
|
88.53 |
|
14 |
[AGAT]14 |
106.32 |
|
11 |
|
82.61 |
|
|
11 |
|
119.25 |
|
|
|
|
|
14 |
|
97.92 |
|
|
|
|
|
15 |
|
94.90 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2 |
9 |
|
111.18 |
|
11 |
|
122.50 |
|
11 |
[ATA]11 |
88.57 |
|
14 |
|
106.48 |
|
14 |
|
91.87 |
|
|
10 |
|
115.21 |
|
12 |
|
126.58 |
|
|
|
|
|
16 |
|
114.42 |
|
15 |
|
94.99 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3 |
9 |
[CTAT]9 |
111.20 |
|
11 |
|
122.46 |
|
11 |
[ATA]11 |
88.60 |
|
15 |
[AGAT]15 |
110.37 |
|
11 |
[ATA]11 |
82.62 |
|
|
|
|
|
|
12 |
|
126.55 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
4 |
7 |
|
103.35 |
|
12 |
|
126.60 |
|
11 |
|
88.60 |
|
14 |
|
106.39 |
|
14 |
|
91.87 |
|
|
9 |
|
111.34 |
|
13 |
|
130.69 |
|
13 |
|
94.78 |
|
15 |
|
110.35 |
|
15 |
|
94.96 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5 |
11 |
|
119.33 |
|
11 |
[AGAT]11 |
122.49 |
|
10 |
|
85.51 |
|
14 |
|
106.41 |
|
11 |
|
82.63 |
|
|
12 |
|
123.39 |
|
|
|
|
|
15 |
|
100.98 |
|
15 |
|
110.39 |
|
14 |
|
91.85 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
6 |
9 |
[CTAT]9 |
111.24 |
|
11 |
|
122.50 |
|
11 |
|
88.58 |
|
14 |
[AGAT]14 |
106.43 |
|
11 |
|
82.62 |
|
|
|
|
|
|
|
|
|
|
14 |
|
97.90 |
|
|
|
|
|
15 |
|
94.93 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
7 |
11 |
[CTAT]11 |
119.26 |
|
11 |
|
122.46 |
|
14 |
[ATA]14 |
97.90 |
|
14 |
[AGAT]14 |
106.38 |
|
14 |
|
91.84 |
|
|
|
|
|
|
12 |
|
126.53 |
|
|
|
|
|
|
|
|
|
15 |
|
94.91 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
8 |
8 |
|
107.27 |
|
12 |
|
126.54 |
|
12 |
|
91.60 |
|
15 |
|
110.30 |
|
11 |
|
82.54 |
|
|
9 |
|
111.27 |
|
|
|
|
|
13 |
|
94.71 |
|
16 |
|
114.29 |
|
15 |
|
94.90 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
9 |
7 |
|
103.31 |
|
12 |
[AGAT]12 |
126.59 |
|
11 |
|
88.57 |
|
14 |
|
106.40 |
|
11 |
|
82.64 |
|
|
10 |
|
115.26 |
|
|
|
|
|
14 |
|
97.93 |
|
15 |
|
110.40 |
|
14 |
|
91.84 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
10 |
10 |
|
115.26 |
|
11 |
|
122.47 |
|
11 |
[ATA]11 |
88.55 |
|
13 |
|
102.43 |
|
11 |
|
82.61 |
|
|
11 |
|
119.27 |
|
12 |
|
126.53 |
|
|
|
|
|
14 |
|
106.38 |
|
15 |
|
94.92 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
11 |
7 |
|
102.72* |
|
12 |
[AGAT]12 |
126.26* |
|
11 |
|
88.14* |
|
14 |
|
106.12* |
|
11 |
|
82.17* |
|
|
10 |
|
114.69* |
|
|
|
|
|
14 |
|
97.39* |
|
15 |
|
110.10* |
|
14 |
|
91.35* |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
12 |
10 |
|
114.69* |
|
11 |
|
122.17* |
|
11 |
[ATA]11 |
88.12* |
|
13 |
|
102.15* |
|
11 |
|
82.18* |
|
|
11 |
|
118.71* |
|
12 |
|
126.32* |
|
|
|
|
|
14 |
|
106.12* |
|
15 |
|
94.45* |
Initial quantification experiments indicated an increase in the apparent DNA concentration the longer the units of SRM2391b remained stored at 4 °C. The DNA concentrations for the SRM 2391b unit stored in-house at -80 °C appear higher than those obtained for the components of SRM 2391b stored at -20 °C. These findings could indicate evaporation occurring, however, since all components were affected a more likely scenario is that the DNA is binding to the walls of the component storage tubes and can slowly return to solution, “soak off,” the tube walls the longer the material is stored at 4 °C.
DNA concentrations of SRM 2391b components 1 – 10 as determined by multiple qPCR analysis runs.
*-80 °C stored unit is also known as “in-house”
**-20 °C stored units: 2 units were obtained from MSD, n=4 is the average of replicates from each unit.
#Data from 11/29/07 replicates where enough material was available due to the use of the units for genotyping prior to the last qPCR analysis.
|
|
-80 °C* |
-20 °C** |
-20 °C** |
-20 °C** |
-20 °C** |
|||||
|
|
n=2 10/30/07 |
n=4 10/25/07 AM |
n=4 10/25/07 PM |
n=4 10/30/07 |
n=0,1, or 2# 11/29/07 |
|||||
|
component |
Avg |
sd |
Avg |
sd |
Avg |
sd |
Avg |
sd |
Avg |
sd |
|
1 |
0.32 |
0.03 |
0.08 |
0.02 |
0.13 |
0.02 |
0.19 |
0.02 |
0.61 |
- |
|
2 |
0.44 |
0.03 |
0.17 |
0.03 |
0.29 |
0.02 |
0.37 |
0.02 |
0.89 |
- |
|
3 |
0.40 |
0.01 |
.017 |
0.04 |
0.27 |
0.06 |
0.41 |
0.05 |
- |
- |
|
4 |
0.32 |
0.02 |
0.21 |
0.06 |
0.31 |
0.03 |
0.31 |
0.01 |
0.49 |
0.04 |
|
5 |
0.43 |
0.01 |
0.34 |
0.08 |
0.43 |
0.10 |
0.49 |
0.03 |
0.70 |
0.02 |
|
6 |
0.39 |
0.00 |
0.24 |
0.03 |
0.30 |
0.04 |
0.43 |
0.14 |
0.72 |
0.03 |
|
7 |
0.40 |
0.02 |
0.16 |
0.04 |
0.26 |
0.08 |
0.24 |
0.09 |
0.56 |
0.04 |
|
8 |
0.70 |
0.03 |
0.42 |
0.03 |
0.51 |
0.09 |
0.67 |
0.09 |
0.87 |
0.03 |
|
9 |
0.59 |
0.19 |
0.31 |
0.07 |
0.42 |
0.06 |
0.56 |
0.07 |
0.88 |
0.00 |
|
10 |
0.48 |
0.09 |
0.33 |
0.08 |
0.40 |
0.04 |
0.51 |
0.07 |
0.87 |
0.02 |
These trends observed mandated additional experiments to elucidate the differences between materials stored at -20 °C versus units and in-house stored units at -80 °C. Two additional SRM 2391b units stored at -20 °C, were labeled 1-20 and 2DI respectivity. Unit 1-20 was at -20 °C while the second unit (2DI) remained exposed to dry ice in the laboratory for two days to simulate shipping. After the two days of storage both units were allowed to equilibrate to laboratory ambient temperature prior to qPCR quantification. For unit one the contents of the odd numbers components (1, 3, 5, 7, and 9) were removed and placed in Teflon containers. Unit two had the contents of the even numbered components (2, 4, 6, 8, and 10) transferred to Teflon containers. These emptied component tubes were refilled with 20 µL of TE-4 buffer and gently rotated for 5 h at ambient temperature prior to storage at 4 °C. After 5 days of 4 °C storage all components were warmed to ambient temperature and quantified with a Quantifiler Human Kit. This process was repeated after an additional 2 days at 4 °C storage and 2 h warm-up at ambient temperature. Material “soaked off” the component tube walls was genotyped with Identifiler. All soaked-off materials gave complete profiles on typing.
Results of this experiment are illustrated in the following Table and Figure.
Data from the “soak-off” experiment units of ng/µL.
|
Initial [DNA] |
Soaked off tubes walls |
||||||||||
|
|
|
|
12/05/07 |
|
12/06/07 |
||||||
|
ID |
Avg |
sd |
|
Avg |
sd |
|
Avg |
sd |
|||
|
T1_1 |
0.10 |
0.02 |
so1_-20_1 |
0.06 |
0.01 |
|
0.08 |
0.02 |
|||
|
1_-20_2 |
0.33 |
0.32 |
|
|
|
|
|
|
|||
|
T1_3 |
0.24 |
0.02 |
so1_-20_3 |
0.11 |
0.02 |
|
0.10 |
0.01 |
|||
|
1_-20_4 |
0.35 |
0.02 |
|
|
|
|
|
|
|||
|
T1_5 |
0.32 |
0.02 |
so1_-20_5 |
0.05 |
0.00 |
|
0.08 |
0.02 |
|||
|
1_-20_6 |
0.49 |
0.06 |
|
|
|
|
|
|
|||
|
T1_7 |
0.16 |
0.01 |
so1_-20_7 |
0.06 |
0.03 |
|
0.09 |
0.02 |
|||
|
1_-20_8 |
0.73 |
0.02 |
|
|
|
|
|
|
|||
|
T1_9 |
0.38 |
0.09 |
so1_-20_9 |
0.13 |
0.00 |
|
0.15 |
0.00 |
|||
|
1_-20_10 |
0.91 |
0.04 |
|
|
|
|
|
|
|||
|
Dry Ice treatment |
|||||||||||
|
2DI_1 |
0.23 |
0.03 |
|
|
|
|
|
|
|||
|
T2_2 |
0.21 |
0.04 |
so2DI_2 |
0.08 |
0.01 |
|
0.11 |
0.01 |
|||
|
2DI_3 |
0.38 |
0.05 |
|
|
|
|
|
|
|||
|
T2_4 |
0.18 |
0.02 |
so2DI_4 |
0.11 |
0.02 |
|
0.13 |
0.01 |
|||
|
2DI_5 |
0.52 |
0.01 |
|
|
|
|
|
|
|||
|
T2_6 |
0.26 |
0.07 |
so2DI_6 |
0.08 |
0.02 |
|
0.17 |
0.00 |
|||
|
2DI_7 |
0.29 |
0.04 |
|
|
|
|
|
|
|||
|
T2_8 |
0.19 |
0.02 |
so2DI_8 |
0.06 |
0.00 |
|
0.09 |
0.00 |
|||
|
2DI_9 |
0.59 |
0.07 |
|
|
|
|
|
|
|||
|
T2_10 |
0.38 |
0.00 |
so2DI_10 |
0.15 |
0.00 |
|
0.17 |
0.01 |
|||
|
|
|
|
|
|
|
|
|
|
|||
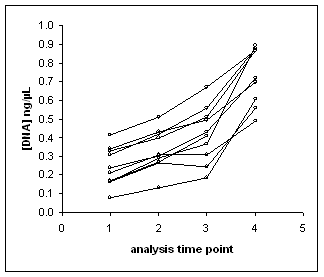
Trends of the analyses of the two MSD units of SRM 2391b. Each line represents an SRM 2391b component and the points along the lines (open circles) correspond to DNA concentrations obtained in this analysis. Several components (2, 8, 9,and 10) were approaching the original nominal DNA concentration of 1 ng/µL at the last time point of 4 days.
MiniFiler D16S539 allelic drop out in Genomic 8 due to a primer binding site mutation
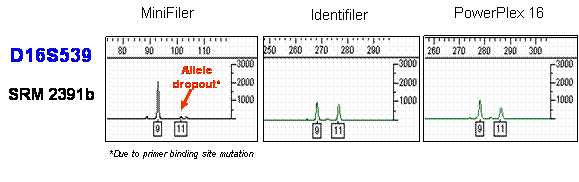
Sequencing results showing MiniFiler D16S539 allelic drop out in Genomic 8

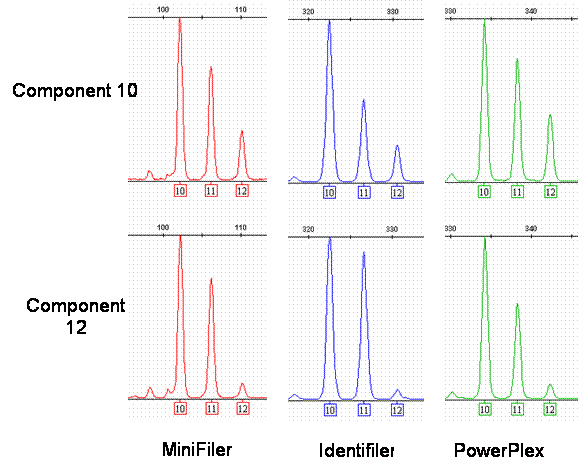
CSF1PO 9948 Tri-Allelic Pattern
A comparison of the tri-allelic profile of components 10 (genomic 9948) and 12 (cell line 9948) at the CFS1PO locus with MiniFiler, Identifiler, and PowerPlex 16